So, I am op !
here is the address of the published project :
h.ttps://smartphone.edgeimpulse.com/classifier.html?publicProjectId=255594
What intrigues me this photo in return ??? Why does it have to be square? How big should the original photo be?
Yes, I just (finally) did it:
h.ttps://smartphone.edgeimpulse.com/classifier.html?publicProjectId=255594
Do you have an idea of the size of the starting photo?
The approach to the problem is not correct. My advice is to start a new project and use the EdgeImpulse FOMO feature to build your model. You do not want to crop out all the Varroa into separate images. You want to draw bounding boxes around each Varroa and train on that labeled data.
FOMO will find objects within an image. FOMO divides an image into meaningful and semantically coherent regions or segments. Instead of classifying the entire image, it aims to assign a label or category to each region within the image.
Your current model is trained to classify the whole image and is called Image Classification and is classifying images into predefined classes or categories, aka, Varroa and ‘non-Varroa’.
The problem with the current approach is:
The current model expects a Varroa to look like this:

When you feed the model something like this:
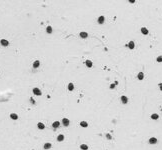
it will classify the image as non-Varroa.
So even a mere human can easily say those 2 images are very different.
ok !
There !
I made a new eim from 38 scans resized 1024x1024 and 38 Yolo.txt files,
it is called: Langes complets avec varroas et non varroas
i got an error in my job :
#!/usr/bin/env python
import cv2
import os
import sys, getopt
import numpy as np
from edge_impulse_linux.image import ImageImpulseRunner
runner = ImageImpulseRunner(“./langes-complets-avec-varroas-et-non-varroas-linux-x86_64-v2.eim”) # C:\Users\berna\Documents\Tensorflow\Edge_Impulse
model_info = runner.init()
print(‘Loaded runner for "’ + model_info[‘project’][‘owner’] + ’ / ’ + model_info[‘project’][‘name’] + ‘"’)
labels = model_info[‘model_parameters’][‘labels’]
img = cv2.imread(“./L61.jpg”)
if img is None:
print(‘Failed to load image’, “./L61.jpg”)
exit(1)
resize img en 1024 x 1024
width = int(1024/img.shape[1])
height = int(1024/img.shape[0])
dsize = (width,height)
img1024 = cv2.resize(img, dsize)
imread returns images in BGR format, so we need to convert to RGB
img = cv2.cvtColor(img1024, cv2.COLOR_BGR2GRAY) # COLOR_BGR2GRAY COLOR_BGR2RGB
get_features_from_image also takes a crop direction arguments in case you don’t have square images
features, cropped = runner.get_features_from_image(img)
res = runner.classify(features)
if “classification” in res[“result”].keys():
print(‘Result (%d ms.) ’ % (res[‘timing’][‘dsp’] + res[‘timing’][‘classification’]), end=’‘)
for label in labels:
score = res[‘result’][‘classification’][label]
print(’%s: %.2f\t’ % (label, score), end=‘’)
print(‘’, flush=True)
elif “bounding_boxes” in res[“result”].keys():
print(‘Found %d bounding boxes (%d ms.)’ % (len(res[“result”][“bounding_boxes”]), res[‘timing’][‘dsp’] + res[‘timing’][‘classification’]))
for bb in res[“result”][“bounding_boxes”]:
print(‘\t%s (%.2f): x=%d y=%d w=%d h=%d’ % (bb[‘label’], bb[‘value’], bb[‘x’], bb[‘y’], bb[‘width’], bb[‘height’]))
cropped = cv2.rectangle(cropped, (bb[‘x’], bb[‘y’]), (bb[‘x’] + bb[‘width’], bb[‘y’] + bb[‘height’]), (255, 0, 0), 1)
the image will be resized and cropped, save a copy of the picture here
so you can see what’s being passed into the classifier
cv2.imwrite(‘debug.jpg’, cv2.cvtColor(cropped, cv2.COLOR_BGR2GRAY)) # COLOR_BGR2GRAY COLOR_BGR2RGB
+++++++++++++ python3 edge_exemple_1_2.py +++++++++++++
ubuntu@vps-16be9656:~/essai_edge$ python3 edge_exemple_1_2.py
Loaded runner for “Berny / Langes complets avec varroas et non varroas”
Traceback (most recent call last):
File “/home/ubuntu/essai_edge/edge_exemple_1_2.py”, line 26, in
features, cropped = runner.get_features_from_image(img)
File “/home/ubuntu/.local/lib/python3.10/site-packages/edge_impulse_linux/image.py”, line 126, in get_features_from_image
g = pixels[ix + 1]
IndexError: list index out of range
ubuntu@vps-16be9656:~/essai_edge$
What can i do ?
Check what is being stored in width and height. I think both are being set to zero.
width = int(1024/3500) = 0
height = int(1024/2500) = 0
just a note; recall that FOMO runs fully convolutionally so can be trained at a resolution that is different to inference time, you just need to make sure that the pixel scale doesn’t change.
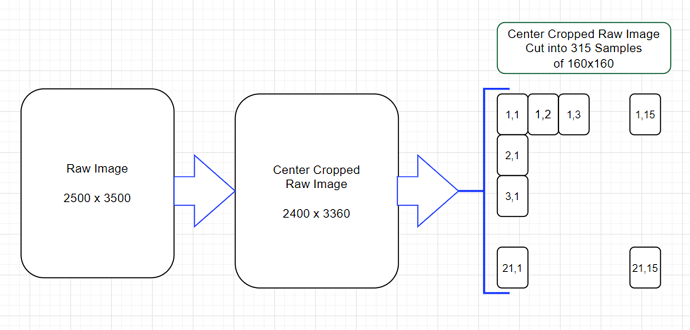
i.e. start with the (2500, 3500) data, sample from that a collection of (160, 160) patches. (160 being the resolution that after the 1/8th reduction is (20, 20) ~= the detection size of interest )
after training this (160, 160) → (20, 20) model it can be run with an input of (2500, 3500) which will output ~= (312, 437). note: if you want to be super strict you’d center crop the input to (2496, 3496) to ensure there’s no boundary artefacts ( i.e. 2496=312*8 )
one main reason to do this is it’ll help the stability of training ( since when we feed in large images we are effectively running with a larger batch size )
cheers,
mat
p.s. have considered doing this patching as a default thing during FOMO training, but also have been working on some large batch size optimisation which hopefully makes the problem redundant.
@MMarcial :
yes, you are right, “width = int(1024/3500) = 0” , it’s a mistake !
So I change and I live the original dimension :
// no resize img en 1024 x 1024
print('Original Dimensions : ',img.shape)
// imread returns images in BGR format, so we need to convert to RGB
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY) # COLOR_BGR2GRAY COLOR_BGR2RGB
but i have the same error :
ubuntu@vps-16be9656:~/essai_edge$ python3 edge_exemple_1_2.py Loaded runner for “Berny / Langes complets avec varroas et non varroas”
Original Dimensions : (2106, 1530, 3)
Traceback (most recent call last):
File “/home/ubuntu/essai_edge/edge_exemple_1_2.py”, line 25, in
features, cropped = runner.get_features_from_image(img)
File “/home/ubuntu/.local/lib/python3.10/site-packages/edge_impulse_linux/image.py”, line 126, in get_features_from_image
g = pixels[ix + 1]
IndexError: list index out of range
@matkelcey :
I will rely on your experience with these numbers and here is my strategy:
-1- I will make a hundred test scans in 2500x3500 from a white sheet on which I will randomly throw a random number of only real varroa mites (between 0 and 20 for example), no “non-varroa”, and notify in Yolo format.
-2- I will follow your path and create an impulse with “image data” = 160 x 160 in “squash”
I should then have an impulse with good results in the “Model testing” phase on 2500x3500 scans.
Then I try on real scans.
What do you think ?
-
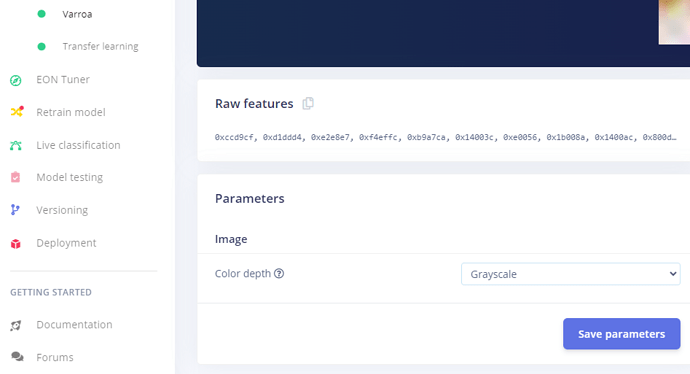
The error in
image.pyis because it thinks the model is RGB. -
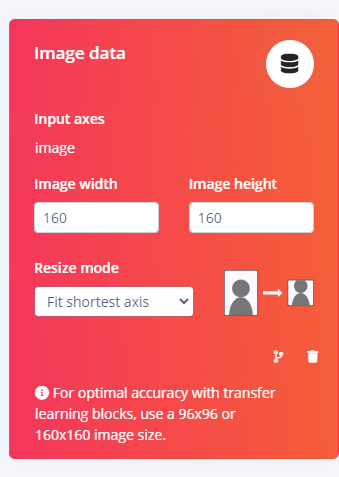
Make sure the Impulse Design is set to Grayscale:
-
Then re-deploy the EIM.
For clarification you are say that we should cut up the raw image for training, correct?
Then does the Impulse design continues to use?:
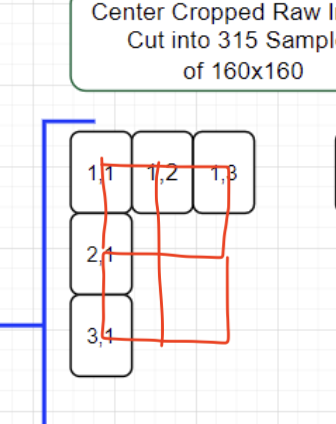
I arbitrarily centered cropped the raw image so I could get an even number of 160x160 Samples (of course there are other ways to handle this since we might be throwing away data).
yeah that’s right. one additional important thing is to do a second pass and collect patches shifted by 1/2 patch size. you want to do this to counter act any boundary effects that are occurring from the patching.
the center cropping of the original and this second pass both drop some of the outer boundary but it’s probably ok to ignore.
sorry, you can’t squash the original image, that’s the same as resizing and would loss almost all the information in the image. i meant what @mmarcial mentioned here ( Find au crop 22x22 in a scan 2500x3500 - #20 by MMarcial ) with respect to cropping out patches
As said before, I made 100 nappies with a number of true varroa varying from 1 to 50 that I sent very easily in edge impulse with the TXT in Yolo.txt format.
I got acceptable test results (42%) and I just published it:
2 Today, 11:23:44 Berny version 100 langes varroas seuls 66% 42% 93 items
this seems to work quite well and i am thinking of trying a version with 500 nappies to see if the performance improves
I will have to study your comments, thank you again
…
If I understand correctly, I have to cut a basic 2500x3500 image into 315 pieces de 160x160 and on each I have to apply the search create with 160x160 “images”, then eliminate the duplicates.
and to reduce the error due to the slicing interfaces I can also do a second slicing shifted from the first by 80.80, which will give me about 13 x 19 new crops to study. 315 + 247 = 562 explorations!!!
…/…
I even believe that to be perfect you have to immerse the 2500x3500 image in a 2560x3520 image that we cut into 352 (i.e.16x22)images of 160 at first then a second time an offset cutout of 315 (i.e.15x21) images, so a total of 667 images of 160x160 largely covering the base image and the interfaces , then apply the create search for 160x160 images and finally remove duplicates.
“Au boulot !!!”
…/…
What I don’t understand is why when I create the pulse, and test it on 2500x3500 images does it work perfectly?? (Model testing)
(I mean without cutting into sub-images of 160x160)
…/…
YES !
I test job passed on a crop of 160x160 !!!
ubuntu@vps-16be9656:~/essai_edge$ python3 edge_exemple_2.py
Loaded runner for “Berny / Langes complets avec varroas et non varroas”
Original Dimensions : (2106, 1530, 3)
Original Dimensions du crop: 1 (160, 160, 3)
Original Dimensions du crop: 3 (160, 160)
Original Dimensions du crop: 2 (160, 160, 3)
Found 12 bounding boxes (71 ms.)
varroa (1.00): x=120 y=0 w=40 h=32
varroa (1.00): x=64 y=40 w=8 h=16
varroa (0.80): x=104 y=48 w=8 h=8
varroa (0.62): x=120 y=56 w=8 h=8
varroa (0.80): x=112 y=88 w=8 h=8
varroa (0.59): x=24 y=96 w=8 h=8
varroa (1.00): x=80 y=104 w=24 h=16
varroa (0.94): x=8 y=112 w=8 h=16
varroa (0.89): x=56 y=120 w=8 h=8
varroa (0.96): x=24 y=128 w=8 h=8
varroa (0.88): x=120 y=136 w=8 h=16
varroa (0.89): x=144 y=136 w=16 h=16
ubuntu@vps-16be9656:~/essai_edge$ ^C
“Thank God, it’s Friday”
Merci beaucoup pour vos explications et votre persévérance
-
As Mr. Matkelcey stated “…FOMO runs fully convolutionally so can be trained at a resolution that is different to inference time, you just need to make sure that the pixel scale doesn’t change.” (I’m still trying to figure out intuitively what this means.)
- Matkelcey assumed you had a detection size of interest of 20x20 pixels. That is why he suggested to train on 160x160 pixels. This choice is an artifact of how the default cutpoint in FOMO works. (Of course the cutpoint in FOMO is user adjustable.)
-
Therefore it is not nessesary to train on 160x160 pixel images if the detection size of interest is not 20x20 pixels.
You basically understand how to chop up the original large image into smaller images for training.
There are many ways to chop up the image and handle the borders. For example, consider row 1 of an image that can handle 4.3 sub-samples. Of course we cannot have a 0.3 sample so we need to decide if we want 4 or 5 sub-samples per row. It comes down to what procedure we will use to place the Varroa in the image. Perhaps it is very expensive to collect this data so we choose to keep all data so we decide on 5 samples per row.
- 1,1 Sample: Starting point.
- 1,2 Sample: Placed adjacent to 1,1 Sample.
- 1,3 Sample: Placed adjacent to 1,2 Sample.
- 1,4 Sample: Placed adjacent to 1,3 Sample.
- 1,5 Sample: Placed adjacent to raw sample right border.
Note sub-samples 1,4 and 1,5 overlap. This is ok.
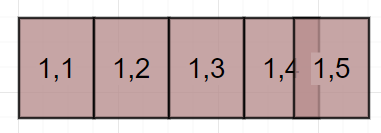
I also do not think you need to delete duplicate bounding boxes that may appear in multiple sub-samples. The same Varroa is in 2 different sub-samples but in different locations so I do not think over-fitting will be an issue. This would make a nice side project to study keeping duplicates versus deleting duplicates.
@matkelcey I fully appreciate the offset-shift to handle errors that might occur when creating sub-samples. In a previous life in another field we called this Grid Registration Error. Perhaps in this case we could call it Boundary Sampling Error (BSE) and the statment would be like, “To counteract BSE we create a second dataset by shifting the original Sample locations by 1/2 in the x and y dimensions”.
and, in very particular cases, shifting by 1/2 the patch size can still cause problems. the absolute best way, least biased, is to random sample patches, but it’s just a tad tricker to orchestrate.
we are looking for a square, whose side is less than 1024, which covers a maximum area and which causes the overlap to be greater than the size of the object sought
2500/3 = 835
3500/4 = 875
object of 22 => 875 + 22 = 897.
I therefore propose 900 to make it simple.
starting at the top left corner flush with the sides of the sheet, ending at the bottom flush with the side of the sheet.
in fact, the minimum seems to me 858.5 or 859 + 22 = 881 maybe less by scratching a little.
900 seems like a good number
…/…
Damn, I put 900 and that’s a bit much for the free version of Edge Impulse:
ERR: Estimated training time (6h 11m 5s) is larger than compute time limit (20m 0s).
Damn, I put 500 :
ERR: Estimated training time (2h 1m 20s) is larger than compute time limit (20m 0s).
Given the constraints of free access, I’m going to make a 272x272 tiling which gives an overlap of 22 10x14 patches to cover a scan of 2500x3500. The other limit is the number of iterations which is aloyrs of 35 max.
What I don’t understand is why when I create the pulse, and test it on 2500x3500 images does it work perfectly?? (Model testing)
(I mean without cutting into sub-images of 160x160 or other)
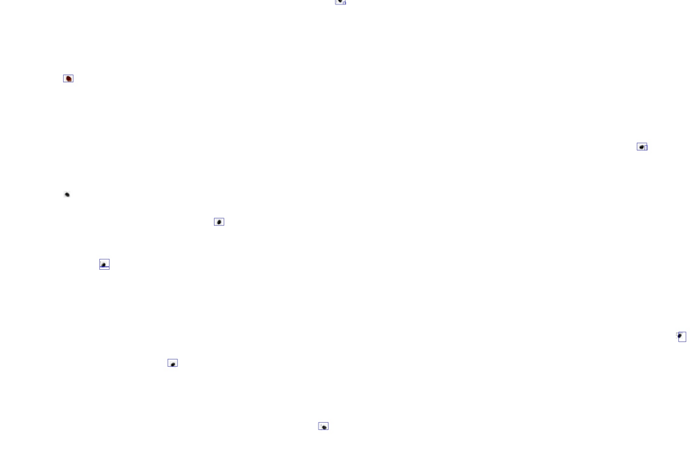
Here I did it with 272x272 patch.
you can see the trace of the 272x272 patch on the scan of 2500x3500
I find my Varroas well on a sheet used for the creation of the AI,
but the result is for the moment a little imprecise on an original sheet with varroas and traces (bee crots, pollen grains, etc.)
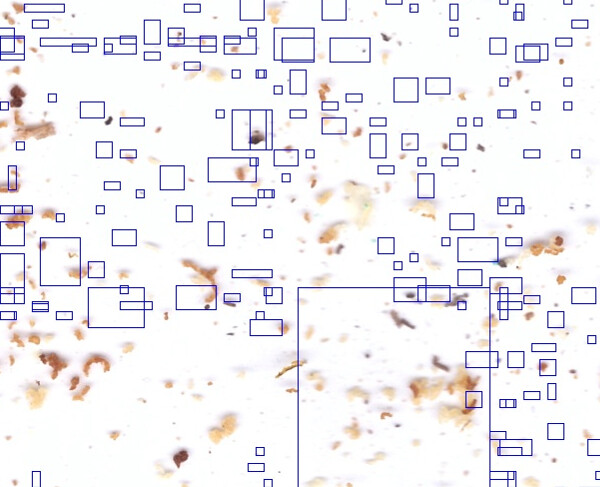
I hesitate:
- either continue with whole leaves in 2500x3500 in Edge Impulse and patches of 272x272
- either only make patches of 272x272 in Edge Impulse
Anyway, the 272x272 patches must be applied to the full leaf of 2500x3500 at the end
What do you think ?
So here it is, I tried the entire A4 sheet and the results are disappointing.
Still, I don’t understand, that’s why when I create the pulse and test it on 2500x3500 footage, does it work perfectly?? (Model test)
On the other hand, the way of focusing on the patches and then applying the patch to the entire surface is more promising. To calculate the size of the patch, I propose to take a value that makes it possible to divide the surface easily and to add to it the size of the object to be searched for in my case I homogenized the size of the A4 scans in 300dpi at 2500x3500 pixels in doing a resize, so I’m going with a patch of 250x250 (I could have taken 250x350) plus the size of the object (here 22x22 pixels) so a patch of 272x272 that I apply 10 times in width and 14 times in length to cover the 2500x3500 pixels with an overlap of 22 in both directions.
So I created 700 images of 272x272 with a 22x22 varroa placed at random and made the impulse whose result is satisfactory: 96% despite the limitation of the processing time to 20 minutes which also limits the number of iterations to 25 d:((
So it is towards a POC that I can orient myself: to prove that it is the right path without being able to reach it.
Frustrating!
Sutrout that as I am not an employee of a company, I cannot even ask for a pricing !