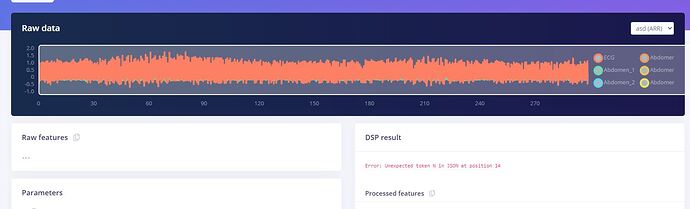
Hello,
I’m working on an ECG dataset, the file (was around 22 MB) was succesfully uploaded on edge impulse but I’m having this problem with the DSP result:
This is the python code used to convert CSV to JSON:
import csv, json, math, hmac, hashlib
jsonFilePath = "prova_ECG.json"
header = None
# keep track of the first row to know the beginning timestamp
first_row = True
begin_ts = 0
next_ts = 0
values = []
HMAC_KEY = "fed53116f20684c067774ebf9e7bcbdc"
# Parse the CSV file
with open("./prova.csv", newline='') as csvfile:
rows = csv.reader(csvfile, delimiter=',')
for row in rows:
if (not header):
header = row
continue
if not begin_ts:
begin_ts = float(row[0])
elif not next_ts:
next_ts = float(row[0])
# skip over timestamp column, and add the rest
values.append([ float(x) for x in row[1:] ])
# empty signature (all zeros). HS256 gives 32 byte signature, and we encode in hex, so we need 64 characters here
emptySignature = ''.join(['0'] * 64)
# This is the Edge Impulse Data Acquisition Format, it has the protected header
data = {
"protected": {
"ver": "v1",
"alg": "none",
"iat": math.floor(begin_ts / 1000) # epoch time, seconds since 1970 (the timestamp earlier was in ms.)
},
"signature": emptySignature,
"payload": {
"device_type": "CSV_IMPORTER",
"interval_ms": next_ts - begin_ts,
"sensors": [ { "name": x, "units": "mV" } for x in header[1:] ],
"values": values
}
}
# encode in JSON
encoded = json.dumps(data)
# sign message
signature = hmac.new(bytes(HMAC_KEY, 'utf-8'), msg = encoded.encode('utf-8'), digestmod = hashlib.sha256).hexdigest()
# set the signature again in the message, and encode again
data['signature'] = signature
encoded = json.dumps(data)
print(encoded)
#Write data to the JSON file
with open(jsonFilePath, "w") as jsonFile:
jsonFile.write(json.dumps(data))
This is the CSV file:
timestamp,ECG,Abdomen_1,Abdomen_2,Abdomen_3,Abdomen_4,Abdomen_5
0,-0.04075297,0.3932546,-0.2895028,0.2495,-0.1919966,0.7912612
0.001,-0.07248314,0.5782515,-0.4120049,0.3582465,-0.2869988,1.111998
…
300.025,-0.03951233,0.007504486,-0.01325134,-0.02374541,-0.03825185,-0.0549877
Anyone could help me with this problem?
Thanks,
Angelo