Hello,
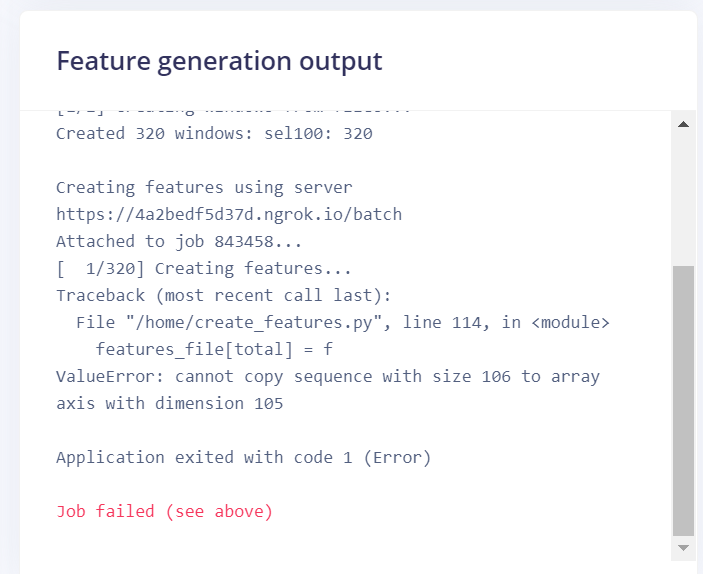
I am trying to extract RR intervals as features from an ECG dataset. Everything seems ok in the first custom block view, but when I try to generate features I got this error:
This is the dsp.py code I used:
import numpy as np
from hrvanalysis import remove_outliers, remove_ectopic_beats, interpolate_nan_values
from hrvanalysis import get_time_domain_features
from hrvanalysis import get_nn_intervals
import json
import peakutils
def smoothing(y, box_pts):
box = np.ones(box_pts) / box_pts
y_smooth = np.convolve(y, box, mode='same')
return y_smooth
# define hrv - da MainCodeYT *Angelo
def hrv(data, fs):
# find peaks
r_peaks = peakutils.indexes(data, thres=0.5, min_dist=0.1)
# RR intervals
RR_intervals = np.diff(r_peaks)/fs
return RR_intervals
def generate_features(draw_graphs, raw_data, axes, sampling_freq, scale_axes, smooth):
# features is a 1D array, reshape so we have a matrix with one raw per axis
raw_data = raw_data.reshape(int(len(raw_data) / len(axes)), len(axes))
features = []
smoothed_graph = {}
# split out the data from all axes
for ax in range(0, len(axes)):
X = []
for ix in range(0, raw_data.shape[0]):
X.append(raw_data[ix][ax])
# X now contains only the current axis
fx = np.array(X)
# first scale the values
fx = fx * scale_axes
# if smoothing is enabled, do that
if (smooth):
fx = smoothing(fx, 5)
# extract features from fx *Angelo
fx = hrv(fx, sampling_freq)
# we save bandwidth by only drawing graphs when needed
if (draw_graphs):
smoothed_graph[axes[ax]] = list(fx)
# we need to return a 1D array again, so flatten here again
for f in fx:
features.append(f)
# draw the graph with time in the window on the Y axis, and the values on the X axes
# note that the 'suggestedYMin/suggestedYMax' names are incorrect, they describe
# the min/max of the X axis
graphs = []
if (draw_graphs):
graphs.append({
'name': 'DSP Graph',
'X': smoothed_graph,
'y': np.linspace(0.0, raw_data.shape[0] * (1 / sampling_freq) * 1000, raw_data.shape[0] + 1).tolist(),
#'y': np.linspace(0.0, 5, 1).tolist(),
'suggestedYMin': -20,
'suggestedYMax': 20
})
return { 'features': features, 'graphs': graphs }
The dsp-server.py code I used is the one given here https://docs.edgeimpulse.com/docs/custom-blocks.
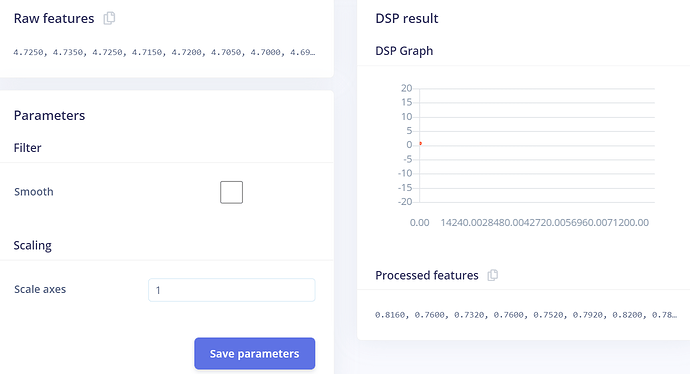
The CSV input file is this:
timestamp,ECG
0,4.725
4,4.735
8,4.725
12,4.715
16,4.72
…
399996,4.765
Anyone could help me with this problem?
Thanks,
Angelo